NeuroJson, founded by Qianqian Fang, is an open-source project which aims to create easy-to-adopt, easy-to-extend, and preferably human-readable data formats to help disseminate and exchange Neuro-imaging data (and scientific data in general). It primarily uses JSON and binary JSON (specifically, UBJSON UBJSON derived Binary JSON format) as the underlying data exchange files.
Table of content
- Caveats of popular tools
- Why NeuroJson
- Testing
UBJson(jdata.jdb)VSNumPy.npyFile size - Discussing Pros and Cons of
.npyand.jdb
Caveats of popular tools
There are just a handful of Python Libraries involved in Medical/Neurological data. NumPy is an essential library that is widely employed in Neuro-imaging data analysis. I have seen discussions about potentially adopting .npy as a standardized way to share Mass Neurological data as seen in this thread.
However, several limitations were also discussed:
.npyonly support a single NumPy array and does not support other metadata or other more complex data records (multiple arrays are only made via multiple files)- no internal (i.e. data-level) compression, only file-level compression
- Even though the file is simple, it still requires a parser to read/write, and such a parser is not widely available in other environments, making it primarily limited to exchanging data among python programs
Why NeuroJson?
Since NeuroJson uses binary-JSON (precisely UBJSON file format), It was able to address most of the Limitations mentioned above ↑. The generated files are universally parsable in nearly all programming environments with existing parsers and support complex hierarchical data with compression. A UBJSON/BJData parser may not necessarily be longer than a .npy parser. For example, the python parser of full specifications only takes about 500 lines of code (including comments), similarly to a JS parser. Generated files from this parser can readily benefit from the large ecosystem of JSON (JSON-schema, JSONPath, JSON-LD, jq, numerous parsers, web-ready, NoSQL DB). Here is a BJson-Parser.
NeuroJson’s team conducted a benchmark test a few months back. The test workloads were two large 2D numerical arrays (node, face to store surface mesh data). They compared the parsing speed of various formats in Python, MATLAB, and JS. The uncompressed UBJData (BMSHraw) reported a loading speed that was nearly as fast as reading a raw binary dump, and internally compressed UBJData (BMSHz) gives the best balance between small file sizes and loading speed,
Results
UBJson(jdata.jdb) VS NumPy.npy File size
A quick comparison of output file sizes with a 1000x1000 unitary diagonal matrix. I wanted to test and compare these formats against each other on my own PC. Here’s the code!
pip install jdata bjdata
import numpy as np
import jdata as jd
x = np.eye(1000) # Creating a large array
y = np.vsplit(x, 10) #Spiliting array in 10 chunks
np.save('eye_10_chunks.npy',y) #Saving as .npy
jd.save(y, 'eye_10_chunks_bjd_raw.jdb') #saving as uncompressed bjd (binary_Json_Data)
jd.save(y, 'eye_10_chunks_bjd_zlib.jdb', {'compression':'zlib'}) #zlib compressed bjd
jd.save(y, 'eye_10_chunks_bjd_lzma.jdb', {'compression':'lzma'}) #Lzma compressed bjd
newy=jd.load('eye_10_chunks_bjd_zlib.jdb') #loading/decoding
newx = np.concatenate(newy) #Regrouping chunks
newx.dtype
Here are the output File sizes in bytes and Loading time in seconds:
eye_10_chunks.npy - 8000120 Bytes/0.177 Seconds
eye_10_chunks_bjd_raw.jdb - 5004297 Bytes/0.719 Seconds
eye_10_chunks_bjd_zlib.jdb 10341 Bytes/1.471 Seconds
eye_10_chunks_bjd_lzma.jdb 2205 Bytes/0.638 Seconds
Discussing Pros and Cons of .npy and .jdb
Pros and Cons of .npy format
Pros:
* Simple ndarrays without additional internal metadata.
* Simple structure lends itself to easy reimplementation whenever needed
* Existing implementations for multiple languages
* .npy files with time: 0.13 seconds, This is by far the fastest method of loading data.
Refer to: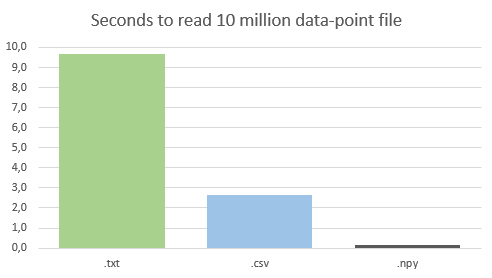 Cons:
* Non-appendable, only possible by using Third-party_packages that comprises processing speed.
Cons:
* Non-appendable, only possible by using Third-party_packages that comprises processing speed.
-
Requires a parser to read/write, and such a parser is not widely available in other environments, making it primarily limited to exchanging data among python programs.
-
No data compression available(Only file), This Leads to big bulky files. For example:

-
Only support a single NumPy array data type and does not support other metadata
Pros and Cons of .jdb format
Pros:
- Appending data is possible, Requires no external package is required
- No external parser is Required,
.jdbgenerated files are universally parsable in most the programming environments - Extremely Compressible, especially using Lzma compressor, it can shrink the data like this:-
a BJData file storing a single eye(10000) array using the ND-array container can be downloaded from here http://neurojson.org/wiki/upload/eye1e4_bjd_raw_ndsyntax.jdb.zip(file size: 1MB with zip, if decompressed, it is ~800MB, as the npy file) - this file was generated from a MatLab encoder but can be loaded using Python (see below Re Robert).Read Thread
Cons:
- Not Optimized for speed, five times slower loading speed than .npy
- The Data needs to be stored in chunks in order to be fully compressible
Conclusion
NeuroJson is emerging as a Major competitor to libraries like Nipy and NumPy in the field of Neuro-Imagery. It is highly compressible and has an established network of JSON/UBJSON files. However, the big challenge is the speed. it is not anywhere near Python’s standard, which presents a big challenge for people working on medical/Neuro-Imaging data Still, I have a positive outlook on NeuroJson that it can become the “Go-To” library for medical-coder and Neuro enthusiasts in the future!

